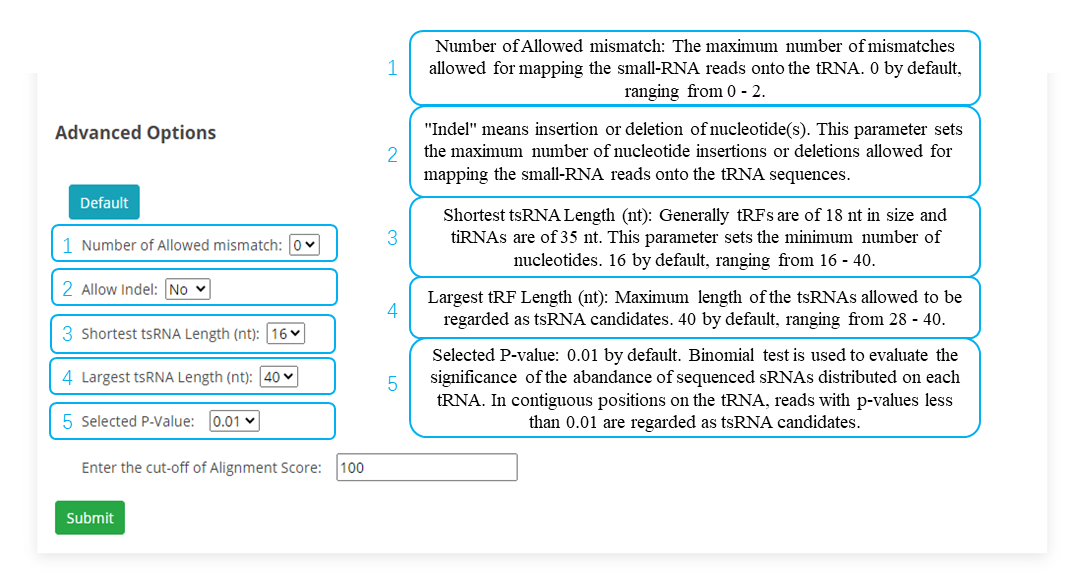
tsRFun Inputs Format
Inputs of tsRFinder and tsRTarget section require users input collapsed FASTA data. The data are recommended to be compressed to increase the uploading speed.
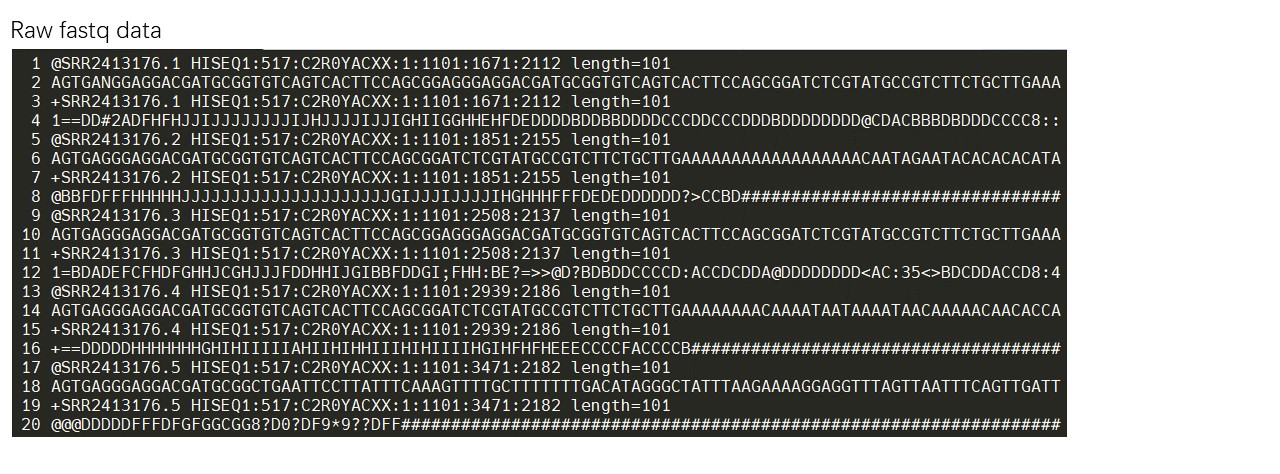
Firstly, users should filter the adapters and low quality reads from raw fastq data. Trim_galore and some other software can trim the adapters and low quality reads at same time.
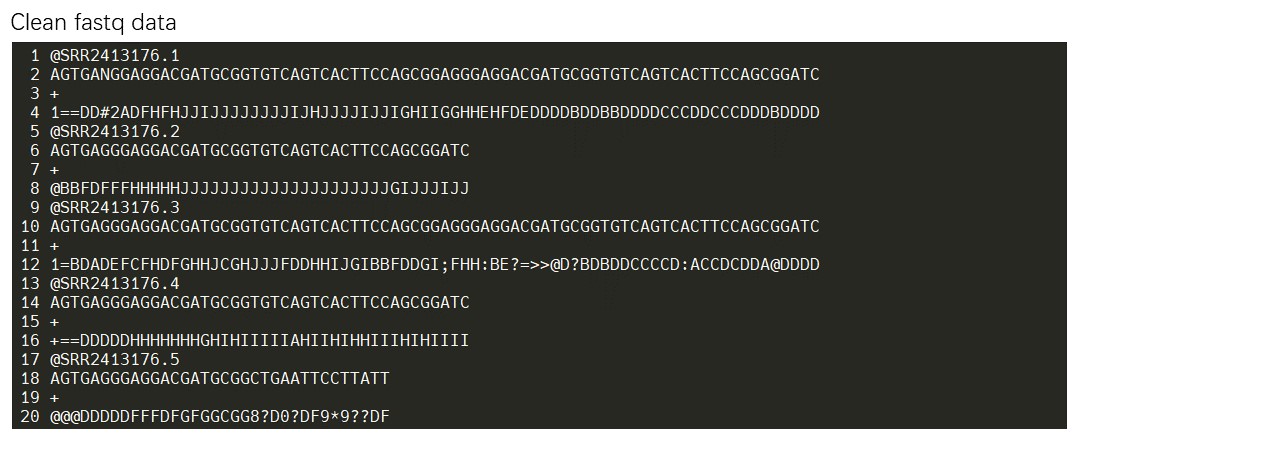
Then, users should collapse the clean fastq reads to fasta reads. The conversion can be done by the following code in Linux.
The collapsed fasta reads are required as input format, and the reads should be named by unique tags (e.g. “>seq_1_x1”. The name includes three parts, first part is “>seq”, the second part “1” represent the unique id, the last “x1” represent the count number of read, and “_” acts as a connector in the tag).