Research Highlights
实验室主要开展生物医药高性能计算研究,结合高性能计算和人工智能技术,跨时空尺度研究生物医药中的重大科学问题。主要研究方向包括
药物智能设计、
蛋白质结构和功能预测、以及
知识驱动的多组学大数据分析,并开发基于“天河二号”的生物医药高性能计算平台。
Back to Top
围绕药物设计全流程,开发了基于AI的全流程算法,包括药物筛选、分子优化、ADMET性质预测、化学合成路线预测等。相关代表性工作包括:
1) 药物智能筛选
• Zheng S#, Y Li#, S Chen, J Xu* and Yuedong Yang*. Predicting Drug Protein Interaction using Quasi-Visual Question Answering System. Nature Machine Intelligence 2020;2(1):134-140. (将蛋白质-药物相互作用问题转化为经典的视觉问答(VQA)问题,启发了分子相互作用研究新范式;荣获2021年世界人工智能大会青年优秀论文奖)
• Wang P, Zheng S, Jiang Y, Li C, Liu J, Wen C, Atanas P, Qian D*, Chen H*, Yuedong Yang*. Structure-aware multi-modal deep learning for drug-protein interactions prediction. J Chem Inf Model 2022; 62(5):1308–1317. (首次在超过4900万个数据点的工业级大数据上进行验证测试)
• Chen P, Ke Y, Lu Y, Du Y, Li J, Yan H, Zhao H, Zhou Y, Yuedong Yang*. DLIGAND2: an improved knowledge-based energy function for protein-ligand interactions using the distance-scaled, finite, ideal-gas reference state. J Cheminform 2019 Aug 7;11(1):52 (基于统计的自由能评估函数)
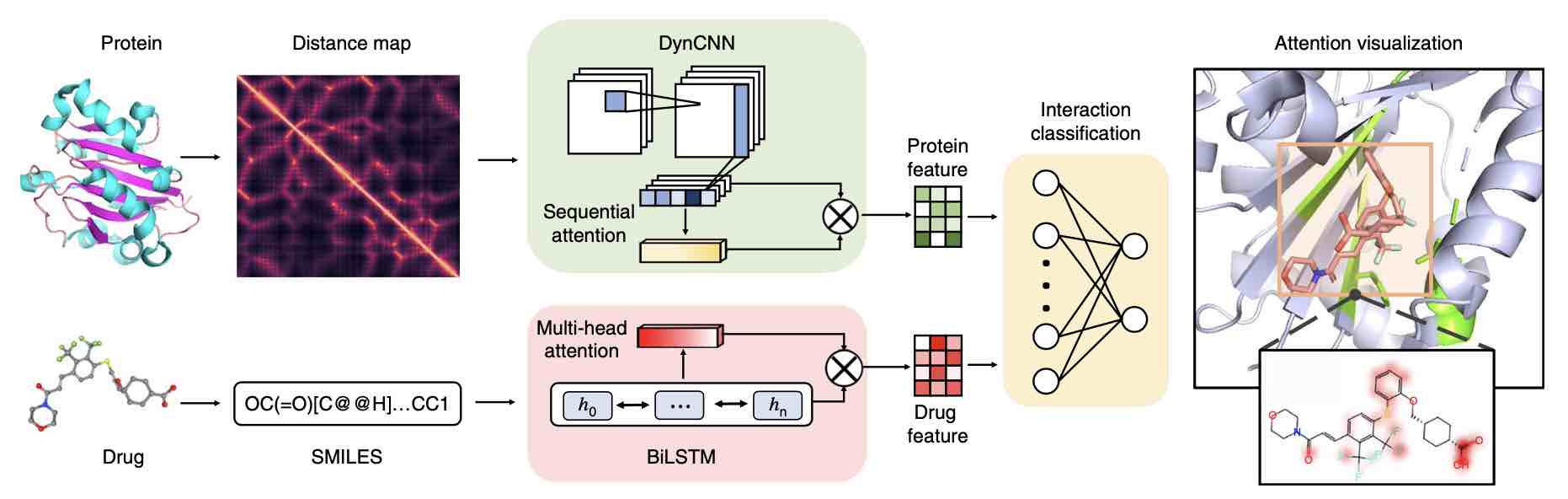
Drug-VQA: Protein-Drug Interaction Prediction
2) 药物性质预测&基础图卷积算法
• Song Y# , S Zheng# , Z Niu , Z Fu , Y Lu and Yuedong Yang*. Communicative Representation Learning on Attributed Molecular Graphs. IJCAI 2020 (AI Top conference, DOI: 10.24963/ijcai.2020/388. (研发了一种新型CMPNN的图卷积模型,在药物性质预测方面有所突破)
• Chen J#, Zheng S#, Yuedong Yang*. Learning Attributed Graph Representation with Communicative Message Passing Transformer. IJCAI 2021. (综合CMPNN和Transformer的新型图卷积框架)
3) 分子优化
• Zheng S, Z Lei, H Ai, H Chen, D Deng*, Yuedong Yang*. Deep Scaffold Hopping with Multi-modal Transformer Neural Networks. J Cheminfo 2021; 13:87.(有效生成维持分子活性的情况下,生成其它方面性能优异的分子)
• Wang J, Zheng S, Chen J, Yuedong Yang*. Meta Learning for Low Resource Molecular Optimization. J Chem Inf Model 2021; (基于元学习的分子优化)
• Zheng S, Rao J, Zhang Z, Xu J*, Yuedong Yang*. Predicting Retrosynthetic Reactions Using Self-Corrected Transformer Neural Networks. J Chem Inf Model. 2020 Jan 27;60(1):47-55.(首次将Transformer用于药物反应路径预测)
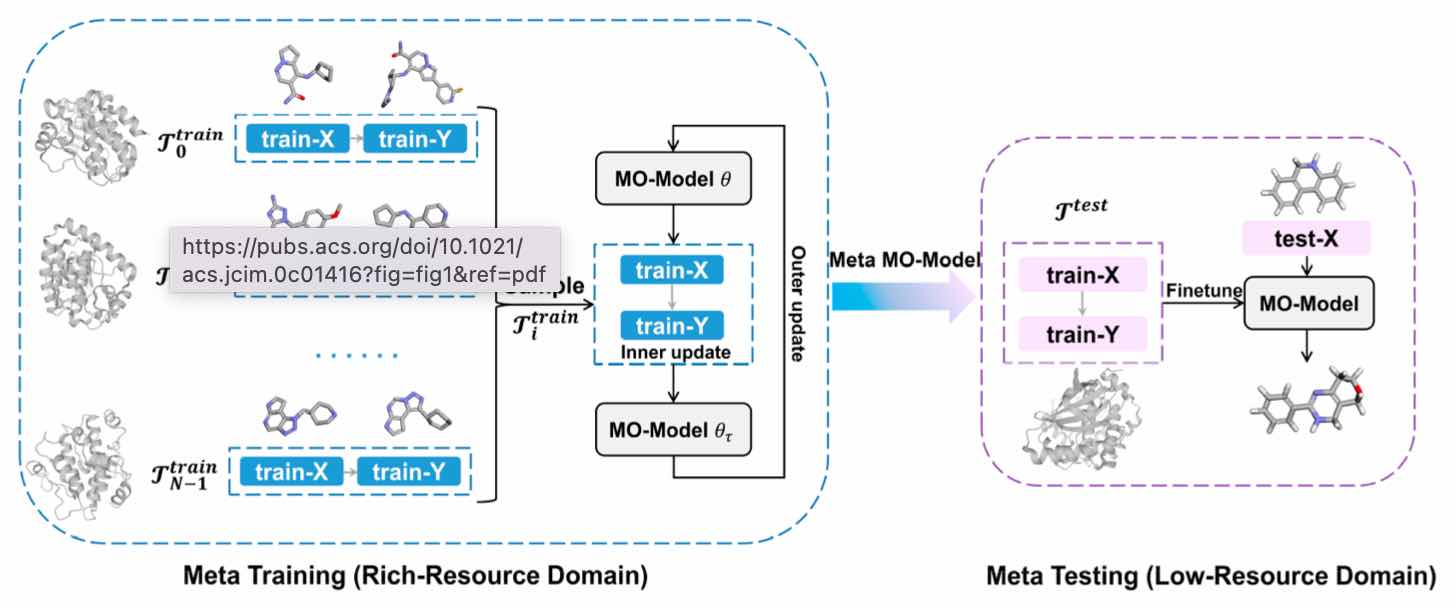
Meta-MO: Meta Learning Model for Molecular Optimization
Back to Top
蛋白质是生物体最重要大分子之一,参与几乎所有的生命活动,准确预测蛋白质三维空间结构和折叠过程被列为21世纪重大科学难题之一。从2013年起,PI就利用深度学习技术开发出蛋白质二级结构预测SPIDER系列,是国际上最早将深度学习用于蛋白质二级结构预测的研究之一,此后不断引入多任务学习、模型的迭代训练等策略,并将结构预测从以前的二级结构离散状态转换为连续数值预测。
1) 蛋白质功能预测
• Yuan Q, Chen S, Rao J, Zheng S, Zhao H, Yuedong Yang*. AlphaFold2-aware protein-DNA binding site prediction using graph transformer. Brief in Bioinfo 2022; bbab564 . (结合Alphafold预测模型的位点预测)
• Yuan Q, Chen J, Zhao H, Zhou Y*, Yuedong Yang*. Structure-aware protein-protein interaction site prediction using deep graph convolutional network. Bioinformatics 2021; btab643. (利用GCN从三维结构预测结合位点)
• Chen J, Zheng S, Zhao H, Yuedong Yang*. Structure-aware Protein Solubility Prediction From Sequence Through Graph Convolutional Network And Predicted Contact Map. J Cheminfo 2021; 13(1):7 (首次利用预测接触图和GCN实现从序列的蛋白质预测)
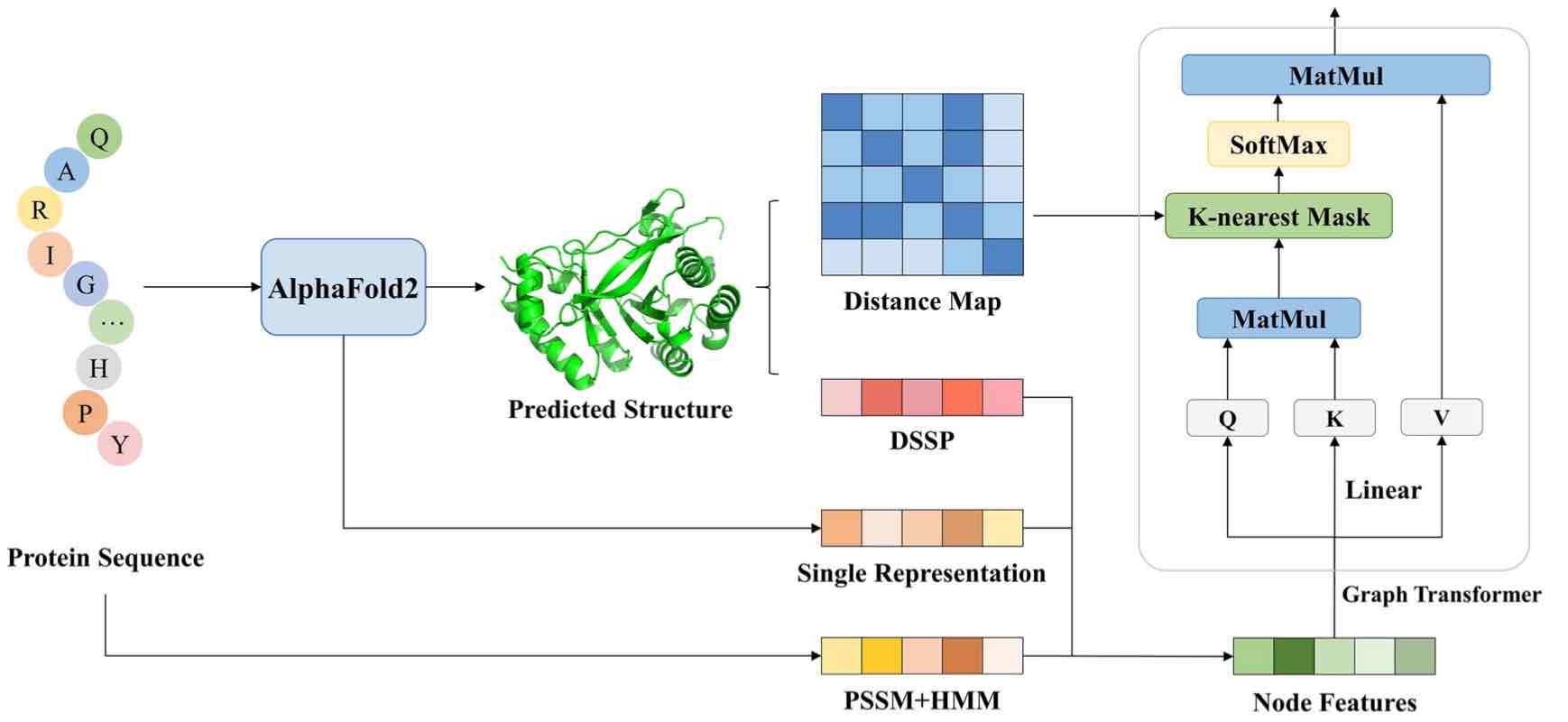
GraphSite: Binding Site Prediction based on Modelled Structures by Alphafold2
2) 蛋白质结构性质预测:
• J Lyons, A Dehzangi, R Heffernan, A Sharmaa, K Paliwal, A Sattar, Y Zhou*, Yuedong Yang*. Predicting backbone Calpha angles and dihedrals from protein sequences by stacked sparse auto-encoder deep neural network. J Comput Chem. 2014; 35(28):2040-6. doi: 10.1002/jcc.23718. (SPIDER,最早的深度学习用于蛋白质连续角度预测)
• Yuedong Yang , J Gao, J Wang, R Heffernan, J Hanson, K Paliwal, and Y Zhou. Sixty-five years of long march in protein secondary structure prediction: the final stretch? Brief in Bioinfo 2018 May 1;19(3):482-494. (SPIDER,二级结构预测)
• Heffernan R, Yuedong Yang*, Paliwal K, Zhou Y*. Capturing Non-Local Interactions by Long Short Term Memory Bidirectional Recurrent Neural Networks for Improving Prediction of Protein Secondary Structure, Backbone Angles, Contact Numbers, and Solvent Accessibility. Bioinformatics. 2017 Sep 15;33(18):2842-2849.
3) 蛋白质三维结构预测
• Chen S, Zhang S, Li X, Liu Y, and Yuedong Yang*. SEGEM: a Fast and Accurate Automatic Protein Backbone Structure Modeling Method for Cryo-EM. BIBM 2021. (基于AI的冷冻电镜结构自动建模;基于该模型,在国家蛋白质科学中心与阿里云联合举办的冷冻电镜复合物结构建模中荣获冠军,并大幅领先其他模型)
• Yuedong Yang, Faraggi E, H Zhao, Zhou Y. Improving protein fold recognition and template-based modeling by employing probabilistic-based matching between predicted one-dimensional structural properties of the query and corresponding native properties of templates. Bioinformatics 2011 Aug 1;27(15):2076-82. (SPARKS,基于模板的三维结构预测)
• Cai Y, Li X, Sun Z, Lu Y, Zhao H, Hanson J, Paliwal K, Litfin T, Zhou Y*, Yuedong Yang*. SPOT-Fold: Fragment-Free Protein Structure Prediction Guided by Predicted Backbone Structure and Contact Map. J Comput Chem. 2020 Mar 30;41(8):745-750. (结合预测性质和分子动力学模拟的从头结构建模)
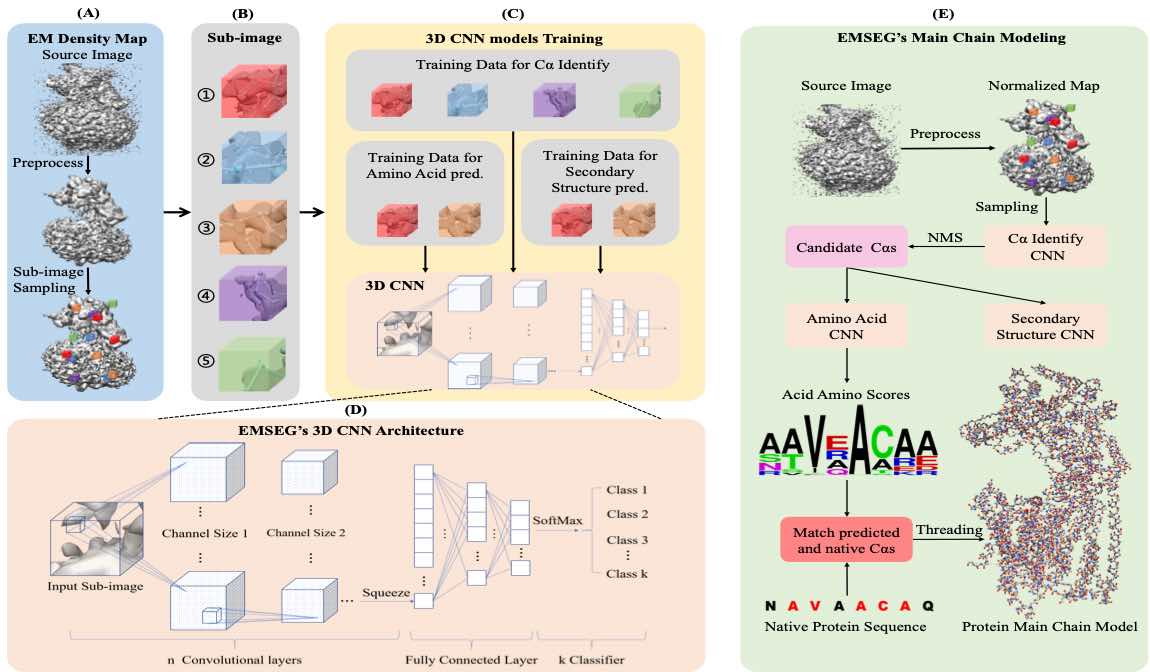
EM-SEG: Automatic Protein Structure Constructing Method for Cryo-EM
Back to Top
随着组学数据多样化和规模化,能从多个时空尺度、不同视角全面阐释生物个体的状态,使得多组学数据分析扮演着越来越重要的角色。然而,多组学的多噪音、高维度、及变量间的复杂关系,需要借助先验知识,才能实现准确的多组学数据分析。
1) 生物医药知识图谱
• Rao J, Zheng S, Mai S, Yuedong Yang*.Communicative Subgraph Representation Learning for Multi-Relational Inductive Drug-Gene Interaction Prediction. IJCAI 2022. (基于子图预测的药物-基因相互作用预测)
• Zheng S#, Rao J#, Song Y, Zhang J, Xiao X, Fang EF, Yuedong Yang*, Niu Z*. PharmKG: A Dedicated Knowledge Graph Benchmark for Biomedical Data Mining. Brief in Bioinfo 2020 (IF=9.0); doi:10.1093/bib/bbaa344. (通过整合OMIM、DrugBank等多个相关公共知识数据库,并从最新文献数据库进行知识抽取,结合人工和算法进行精细的数据清洗和实体性质对齐,最终包括基因、药物和疾病三大类共8000余种实体、和它们之间29类50多万个相互关系)
• Mai S#, Zheng S#, Yuedong Yang*, Hu H*. Communicative Message Passing for Inductive Relation Reasoning. AAAI 2021(基于归纳的知识推导CoMPILE算法)
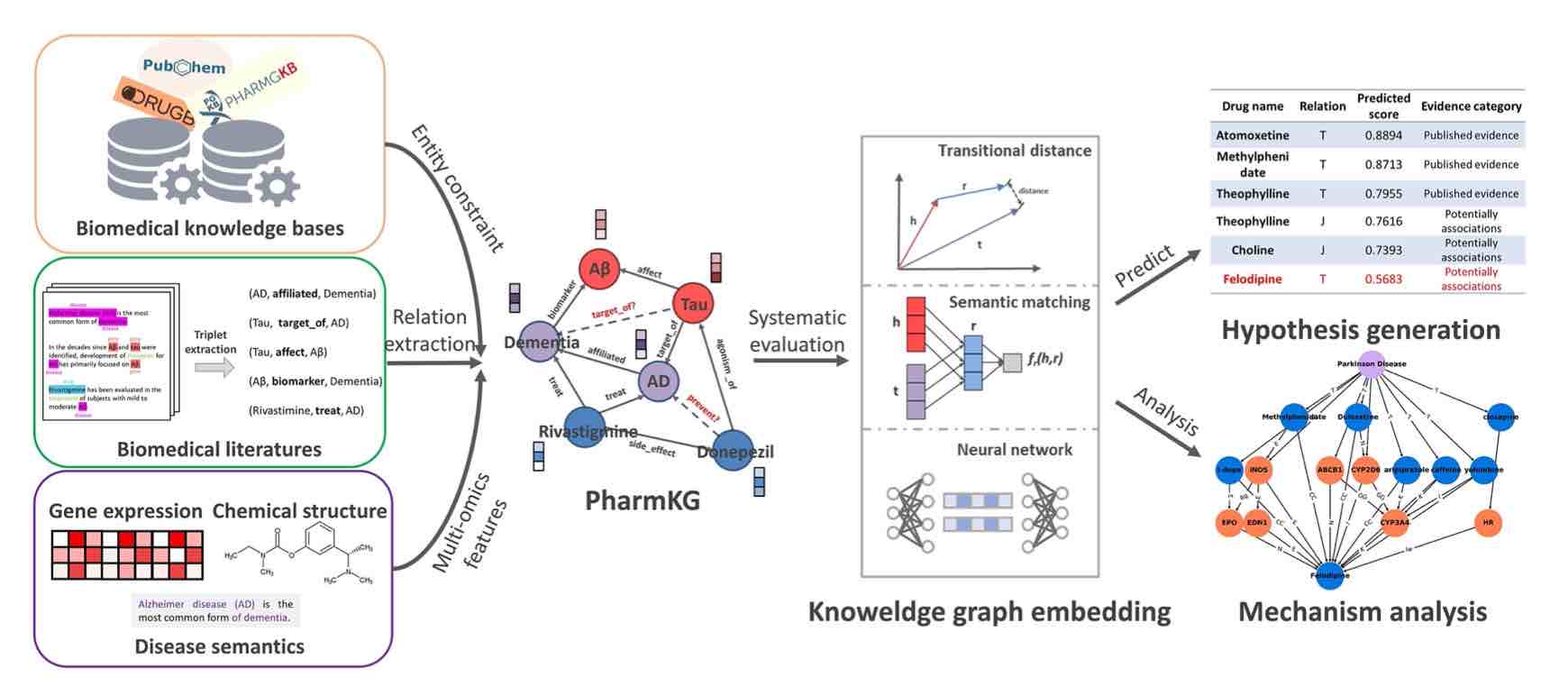
PharmKG: Biomedical Knowledge Graph
2) 单细胞数据分析
• Zeng Y, Zhou X, Pan Z, Lu Y*, Yuedong Yang*. A Robust and Scalable Graph Neural Network for Accurate Single Cell Classification. Brief in Bioinfo 2022; bbab570. (结合PageRank算法的超快速单细胞分类)
• Zeng Y, Wei Z, Zhong F, Pan Z, Lu Y, Yuedong Yang*. A Parameter-free Deep Embedded Clustering Method for Single-cell RNA-seq Data. Brief Bioinfo 2022 (无参单细胞聚类)
• Zhou X, Chai H, Zeng Y, Zhao H, Yuedong Yang*. scAdapt: Virtual adversarial domain adaptation network for single cell RNA-seq data classification across platforms and species. Brief in Bioinfo 2021, bbab281 & RECOMB 2021. (基于域对齐的批次消除)
• Rao J, Zhou X, Lu Y, Zhao H, Yuedong Yang*. Imputing Single-cell RNA-seq data by combining Graph Convolution and Autoencoder Neural Networks. iScience 2021; 24(5):102393 (第一个基于图卷积的数据补齐)
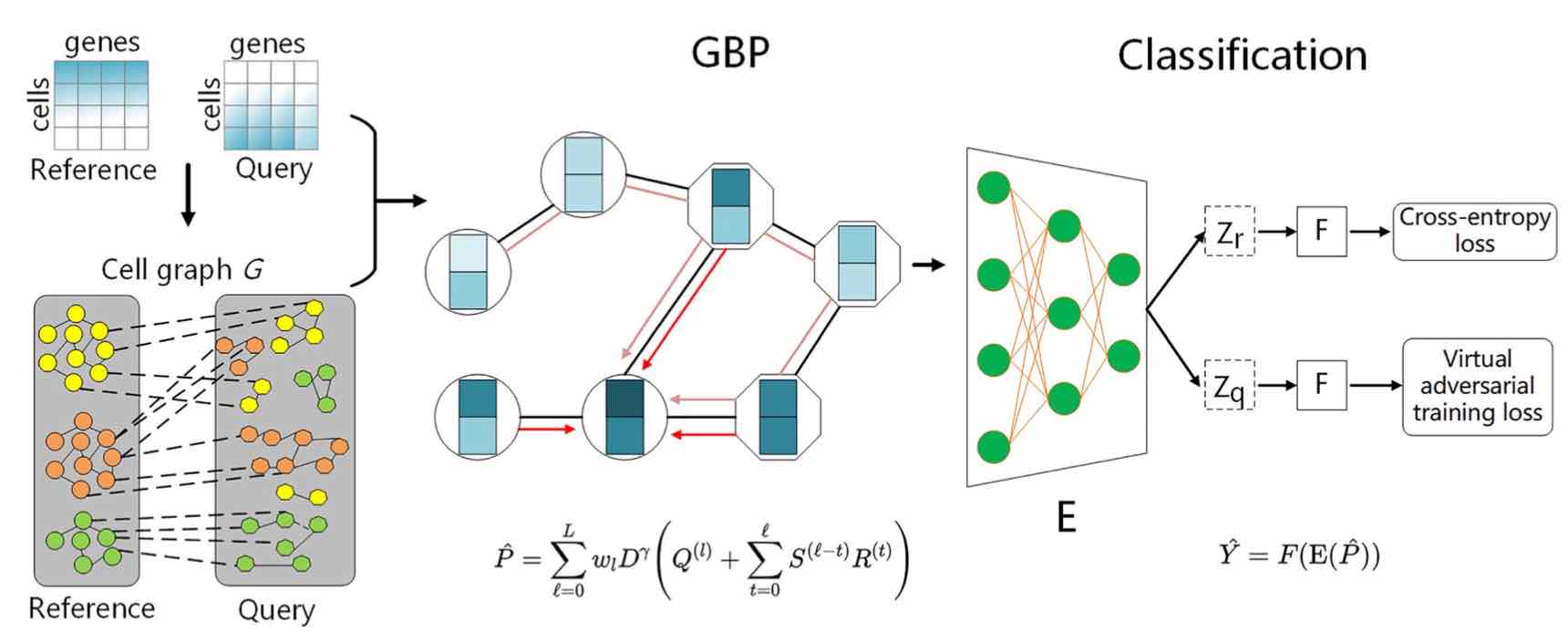
GraphCS: Scalable Single Cell Classification Applicable to Huge Datasets
3) 疾病多组学数据分析
• Chai H, Zhou X, Zhang Z, Rao J, Zhao H, Yuedong Yang*. Integrating multi-omics data with deep learning for predicting cancer prognosis. Comput Biol Med 2021; 134:104481. (多组学癌症预后分析)
• Huang Z, H Chai, R Wang, H Wang, Yuedong Yang*, H Wu*. Integration of Patch Features through Self-Supervised Learning and Transformer for Survival Analysis on Whole Slide Images. MICCAI 2021. (基于病理图像的存活预测)
• Song Y, Zheng S, Li L, Zhang X, Zhang X, Huang Z, Chen J, Zhao H, Jie Y, Wang R, Chong Y*, Shen J*, Zha Y*, Yuedong Yang*. Deep learning Enables Accurate Diagnosis of Novel Coronavirus (COVID-19) with CT images. IEEE TCBB 2021;18(6):2775-2780. (第一个COVID-19的医疗影像深度预测模型)
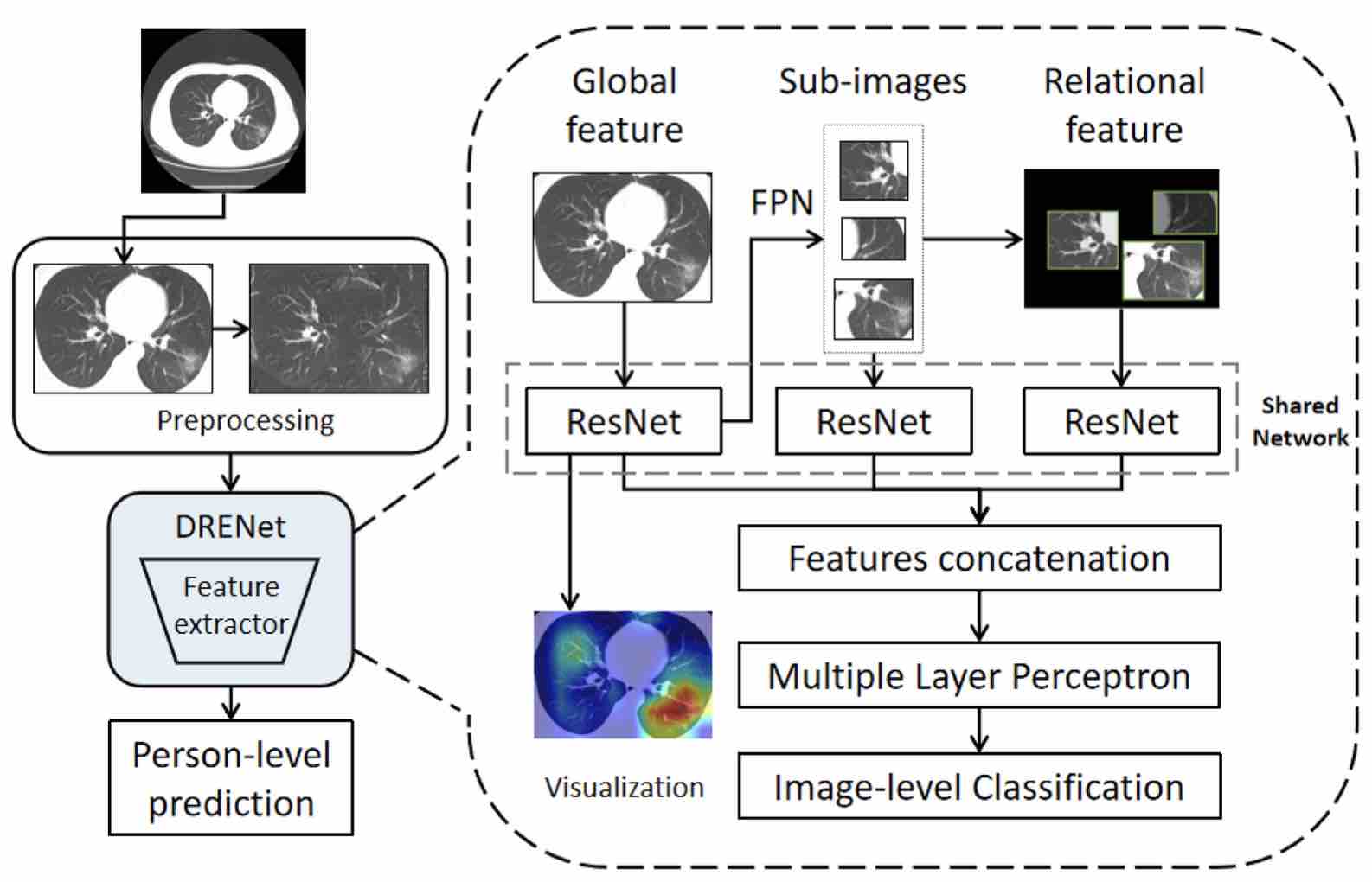
COVID19-net: COVID-19 Diagnosis Method based on CT images






